|
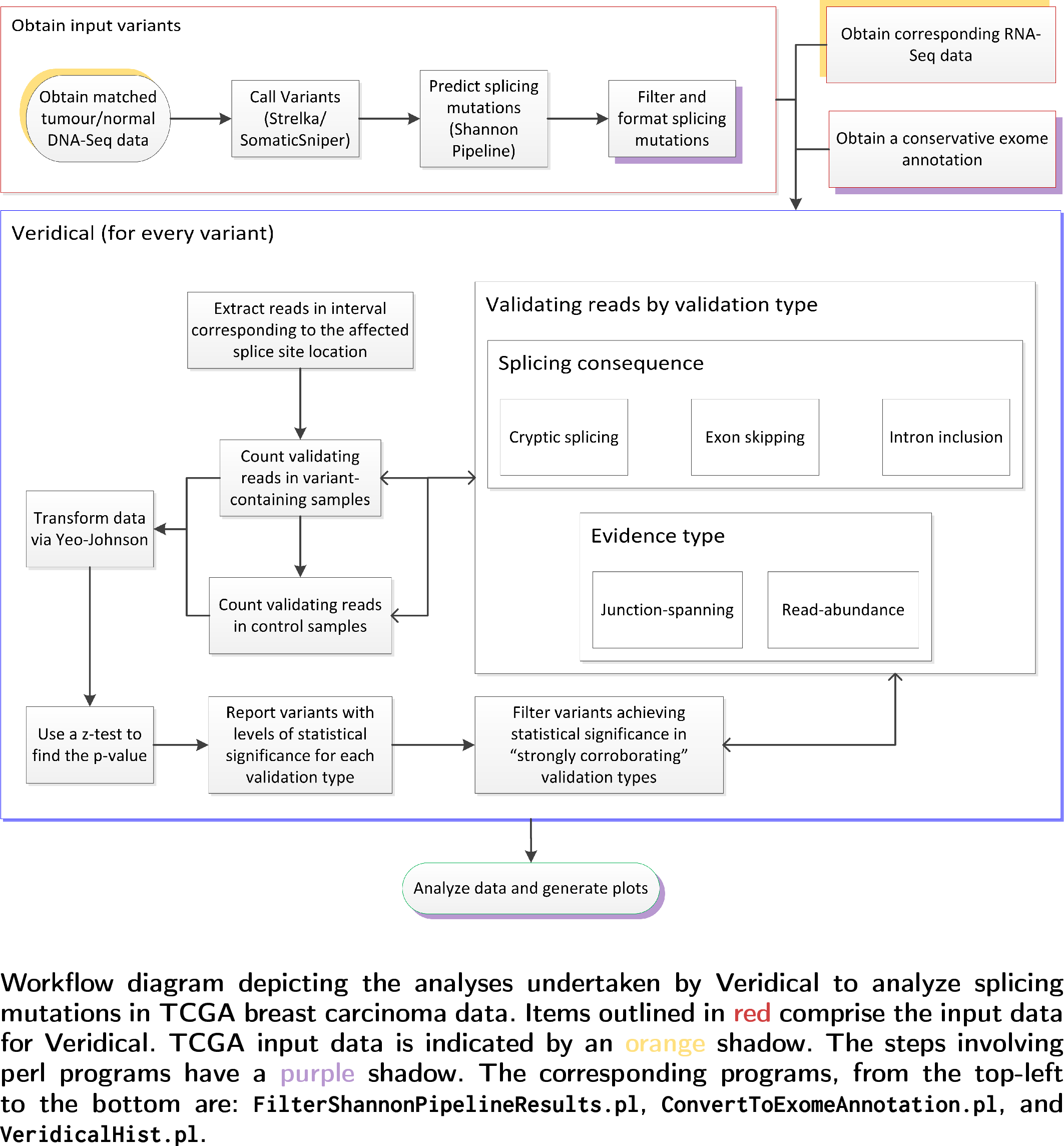
Veridical is companion software to the article "Validation of predicted mRNA splicing mutations using high-throughput transcriptome data", Viner et al. published in F1000research. Veridical provides an in silico method for the automatic validation of DNA sequencing variants that alter mRNA splicing. Veridical performs statistically valid comparisons of the normalized read counts of abnormal RNA species in mutant versus non-mutant tissues. Veridical uses output from the Shannon human splicing pipeline as input. Data from other souces can certainly be used as well, however it must be ordered in such a way to match Shannon pipeline output formatting.
This website is depreciated and is now a part of MutationForecaster. How to cite Veridical: Viner C, Dorman SN, Shirley BC and Rogan PK (2014) Validation of predicted mRNA splicing mutations using high-throughput transcriptome data [v2; ref status: indexed, http://f1000r.es/378] F1000Research 2014, 3:8 (doi: 10.12688/f1000research.3-8.v2) |